The areas of Graph Theory, Link Analysis and Social Network Analysis all hinge on similar underlying concepts. I find this area fascinating in respects to data science. One issue in this area is the large number of very diverse technologies. In many ways a more limited set of tools would free you to think about the problem at hand. If you have no constraints there are probably to many choices bogging you down.
If you need a graph database there are lots of options:
There are even different options for query languages such as Cypher and Gremlin and even the whole Tinkerpop stack to do all sorts of tasks.
For tools to do real analytic work there are different options. I think this consists of two main types.
Graph Processing engines
Graph Analytic libraries
As well as libraries in most general purpose languages
Once you do your analysis you need to visualize the data. There are tons of tools to visualize networks that offer point and click functionality:
If you need to make your own plots programmatically javascript has tons of tools:
What about file formats, there are tons here as well depending on what tool generates the data, can the data change over time, is it directed, can nodes have attributes, can edges? To many choices. Here is actually a site of the options you can use inside of Gephi. I am actually getting overwhelmed with the shear number of different types of tools, much less the specific tools in that domain.
I want to try to walk through a way that you can utilize some of the best methods and tools and have them all communicate with each other.
Getting and Loading the Data
The first thing to address is file formats for graphs. This is really about storing graphs which graph databases would come to mind first but that is a bigger topic. One that allows lots of flexibility in the types of graphs you describe is the gexf format. This was created by Gephi. It may be a little more complicated that some of the simpler methods but it adds a lot of flexibility. As an example the amazing site has gexf files related to movies. I thought it would be very fitting to use The Social Network but it was not there so we have to settle for Pulp Fiction.
Once we have this file we can use the rgexf package to load the data into R.
require(rgexf)
require(httr)
options(stringsAsFactors = F)
# Get the gexf file from the site an place it into a file
gex <- as.character(GET('http://media.moviegalaxies.com/gexf/660.gexf'))
cat(gex, file = 'movie.gexf')
# Read it in with the gexf reader
pulp <- read.gexf('movie.gexf')
# Investigate file
class(pulp)## [1] "gexf"summary(pulp)## GEXF graph object## $`N of nodes`
## [1] 38
##
## $`N of edges`
## [1] 102
##
## $`Node Attrs`
## NULL
##
## $`Edge Attrs`
## NULL# plot(pulp)
I am not sure how to go about including the plot here. This plot is okay, it is using sigmajs, not as great as what Gephi could have done but it is in the browser over stuck in Gephi. Once we have this graph though what do we do with it. Usually we want to know something about the structure of the network. Who is the most central person in the network, does it have any interesting features, does it follow a power law distribution. The file type and the plot have no relevance here, technically I guess you could use the plot to get at these with the old pencil and paper method. We need to turn our network from its gexf form into something more capable for analysis.
Lets try igraph. The gexf package comes with a function to transform gexf to igraph. There is also the reverse to go from igraph to a gexf which can then be consumed by Gephi.
library(igraph)
# Transform to igraph class
ipulp <- gexf.to.igraph(pulp)
plot(ipulp)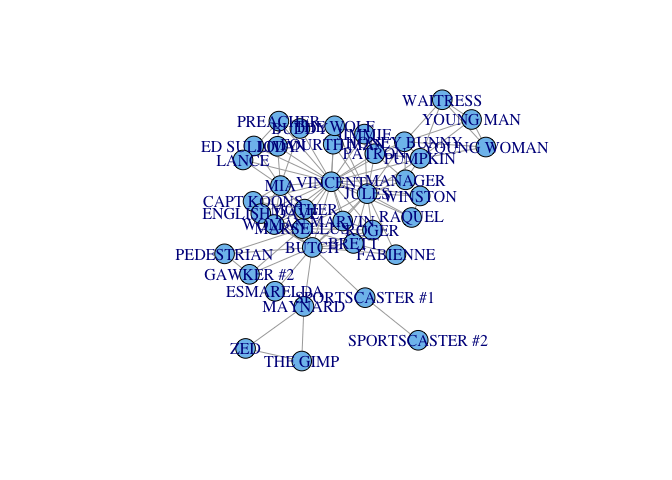
The plot looks pretty bad but we were interested in analysis over aesthetics.
# Analysis at the individual level
scores <- data.frame(alpha = alpha.centrality(ipulp),
authority = authority.score(ipulp)[[1]],
closeness = closeness(ipulp))
head(scores)## alpha authority closeness
## BRETT 1 0.2136 0.01250
## BUDDY 1 0.0790 0.01075
## BUTCH 2 0.3828 0.01429
## CAPT KOONS 3 0.1124 0.01250
## ED SULLIVAN 1 0.0790 0.01075
## ENGLISH DAVE 3 0.1763 0.01053# Analysis at the network level
cliques(ipulp, min = 6, max = 7)## [[1]]
## [1] 1 3 14 17 18 27
##
## [[2]]
## [1] 1 3 14 17 18 32
##
## [[3]]
## [1] 1 3 14 17 27 32
##
## [[4]]
## [1] 1 3 14 18 27 32
##
## [[5]]
## [1] 1 3 17 18 27 32
##
## [[6]]
## [1] 1 14 17 18 27 32
##
## [[7]]
## [1] 3 4 20 21 32 35
##
## [[8]]
## [1] 3 14 17 18 27 32
##
## [[9]]
## [1] 11 14 16 22 25 32
##
## [[10]]
## [1] 1 3 14 17 18 27 32Plotting with D3
Now that we did the math thing and know everything there is to know about this network we want to present our findings. The plot from igraph was pretty rough, the plot from gexf was good but only offered us the ability to zoom in and out and gave us some over capability. We need to give people the ability to interact with the presentation. What we really need here some d3. There is always a package to help us, here it comes in the form of d3network. There is a pretty good tutorial on the website.
We need a way to transform our igraph into the appropriate data structure for d3. The following function takes an igraph class and returns the data frame to send to d3Network plots.
igraph_2_d3 <- function(igr) {
Source <- c()
Target <- c()
for (i in seq(length(E(igr)))) {
e <- get.edge(igr, i)
x <- V(igr)
s <- if(is.null(x[e[1]]$name)) x[e[1]] else x[e[1]]$name
t <- if(is.null(x[e[2]]$name)) x[e[2]] else x[e[2]]$name
Source <- c(Source, s)
Target <- c(Target, t)
}
data.frame(Source, Target)
}
library(d3Network)Now we can just use one of the network plots provided by the package and we get an interactive network that is quite fun to play with.
d3SimpleNetwork(igraph_2_d3(ipulp))We can add more details by changing the the size and color of nodes and edges to have certain features from out analysis, for instance size the nodes by there closeness and color them to differentiate the 7-clique from the rest of the network.
If you are using RStudio you can use this function to make the plot appear in the viewer, which is very useful having everything in one IDE.
d3plot <- function(network, h = 300, w = 700) {
# Create temporary html file
htmlFile <- tempfile(fileext=".html")
if(is.igraph(network)) network <- igraph_2_d3(network)
# Write d3 network viz to html file
d3SimpleNetwork(network, height = h, width = w, file = htmlFile)
# (code to write some content to the file)
rstudio::viewer(htmlFile)
}
d3plot(barabasi.game(20))Persisting with Neo4j
I think my next step would be to persist this data to a graph database. I am going to use Neo4j as the database. Once you download and unzip you just need to run the following at the command line.
bin/neo4j startTo persist the data we need to be able to talk to the database. I looked around and found a few older examples but none actually worked. I think you could use rcurl but I am just going to write my own using system commands as a first go.
library(bitops)
library(RCurl)
library(rjson)
# Put data into format for neo4j
neo_pulp <- igraph_2_d3(ipulp)
head(neo_pulp)## Source Target
## 1 BRETT MARSELLUS
## 2 BRETT MARVIN
## 3 BRETT ROGER
## 4 BRETT VINCENT
## 5 BUDDY MIA
## 6 BUDDY VINCENT# List of all nodes
nodes <- unique(c(neo_pulp$Source, neo_pulp$Target))
head(nodes)## [1] "BRETT" "BUDDY" "BUTCH" "FABIENNE" "FOURTH MAN"
## [6] "GAWKER #2"# Create some common strings.
neoA <- 'Accept:application/json'
neoB <- 'http://localhost:7474/db/data/node/'
neoC <- 'Content-Type:application/json'
# Common helper functions.
qy <- function(x) fromJSON(paste(system(x, intern = T), collapse = ' '))
prep <- function(lst) paste("'", toJSON(lst), "'", sep = '')
# Function to write a node to a database.
create_node <- function(at) {
x <- qy(paste('curl -H ', neoA, ' -H ', neoC, ' -X POST -d ',
prep(at), ' ', neoB, sep = ''))$self
as.integer(gsub(neoB, '', x))
}
# Function to pull node from database.
get_node <- function(id) {
stopifnot(is.numeric(id))
qy(paste('curl -H ', neoA, ' ', neoB, id, sep = ''))$data
}
# Add node and check it exists.
lookup <- create_node(list(name = nodes[1]))
get_node(lookup)## $name
## [1] "BRETT"# Need to create a lookup table, nodes are key: value
nodes_lk <- c()
# Now we just need to loop through all nodes and add them to the database.
for (i in nodes) {
nodes_lk <- c(nodes_lk, create_node(list(name = i)))
}
# Check the lookup table
get_node(nodes_lk[10])## $name
## [1] "JULES"# You can query the database with the following query.
# MATCH (n) OPTIONAL MATCH (n)-[r]-() DELETE n,r;
# Function to create edges between two nodes.
create_edge <- function(to, from) {
from <- nodes_lk[which(nodes == from)]
to <- nodes_lk[which(nodes == to)]
x <- system(paste('curl -H ', neoA, ' -H ', neoC, ' -X POST -d ',
'\'{"to": "', neoB, to, '", "type": "KNOWS"}\' ', neoB, from,
'/relationships', sep = ''), intern = T)
}
# Add each edge to the database
for (i in 1:nrow(neo_pulp)) {
create_edge(neo_pulp$Target[i], neo_pulp$Source[i])
}This is what the console looks like in the Neo4j dashboard.
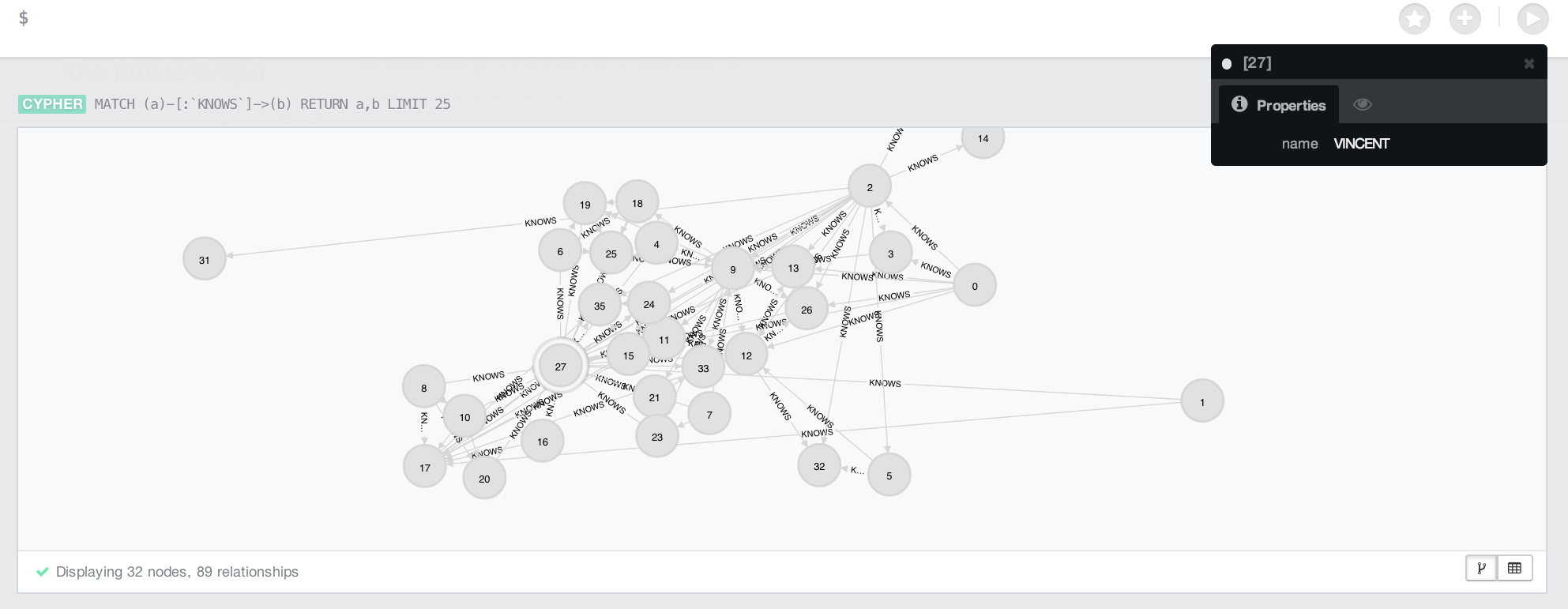
Conclusion
I think I may have actually gone in a very strange order. I started with data from Gephi and did some work to get it into R. After doing some analysis with I plot the data in D3. Then put the data in a graph database. Usually you start with data in a database then do the analysis and then one of the either presentation graphics in Gephi or interactive graphics in D3. It does do what I had hoped in making a bunch of desperate functionality together.
Another technology that I have not worked with here are the large scale graph processing frameworks like Graphx. I have managed to get spark setup so that I can try Graphx but there seems to be a lot more to learn before interacting with it.